Plot2Map is an R package for comparing forest plot data to biomass maps, with a focus on Above Ground Biomass (AGB) estimation and validation.
Installation
You can install Plot2Map from GitHub:
# install.packages("devtools")
devtools::install_github("aTnT/Plot2Map")What Plot2Map can be used for
- Pre-processing various types of plot data (point data, polygons, tree-level measurements, lidar)
- Temporal adjustment of plot data to match map epoch
- Estimation of measurement and sampling errors
- Validation of global AGB maps
- Support for custom forest masks
- Aggregation of results at different spatial scales
Usage
Plot data pre-processing
Here’s a basic workflow using Plot2Map. We’ll be using the plots dataset provided with the package.
library(Plot2Map)
# Load sample plot data. In a real application, this would be your own data
data(plots)
head(plots)
# PLOT_ID POINT_X POINT_Y AGB_T_HA AVG_YEAR SIZE_HA
# 1 AFR7 24.53906 0.9925556 407.60 2011.5 1
# 2 AFR7 24.52981 0.8063056 291.20 2011.5 1
# 3 AFR7 24.52256 0.8274167 366.00 2011.5 1
# 4 AFR10 12.74900 6.2112000 24.90 2007.0 1
# 5 AUS1 145.62743 -37.5924562 699.46 2014.0 1
# 6 AUS1 115.98288 -34.5775977 595.41 2012.0 1We now preprocess the sampled plot data. The first step is to remove plots that have been deforested based on the Global Forest Change (GFC) dataset (Hansen et al., 2013). We use Deforested() that processes each plot, downloads the necessary GFC forest loss tiles, and determines if the plot has been deforested beyond a set deforestation threshold or if the deforestation occurred before or during the specified map year. In the example below we use the default 5% deforestation threshold, and we will validate an AGB map from 2022:
# To reduce data download needs we will only use plots within the "EU2" plot id region
# Keep in mind that this still requires a significant amount of data to be downloaded
preprocessed_plots <- Deforested(plots[plots$PLOT_ID == "EU2", ] , map_year = 2022)
# (...)
# Hansen_GFC-2023-v1.11_lossyear_40N_010W.tif already exists and verified - skipping
# Processing row 5328 with PLOT_ID EU2 and buffered area 0.19 ha ...
#
# Hansen_GFC-2023-v1.11_lossyear_40N_010W.tif already exists and verified - skipping
# Deforestation detected in 100% of the buffered area in plot row 5328 with PLOT_ID EU2.
# The earliest measured year of deforestation is 2016.
#
# Removed 354 plot(s) that have > 5 % deforested area before/during the 2022 map year.
print(preprocessed_plots)
# $non_deforested_plots
# Simple feature collection with 4974 features and 4 fields
# Geometry type: POINT
# Dimension: XY
# Bounding box: xmin: -7.451944 ymin: 36.06354 xmax: 3.24262 ymax: 43.45025
# Geodetic CRS: WGS 84
# First 10 features:
# PLOT_ID AGB_T_HA AVG_YEAR SIZE_HA geometry
# 1 EU2 45.90801 2002 0.196 POINT (-6.740698 38.14153)
# 2 EU2 51.24531 2006 0.196 POINT (-5.06974 37.89271)
# 3 EU2 69.61214 2004 0.196 POINT (-2.326055 38.2154)
# 4 EU2 58.50748 2002 0.196 POINT (-4.500239 40.56834)
# 5 EU2 130.73962 2000 0.196 POINT (-4.524572 43.13481)
# 6 EU2 16.50173 2005 0.196 POINT (-0.9116234 42.72071)
# 7 EU2 25.77160 2006 0.196 POINT (-5.17236 38.28734)
# 8 EU2 77.99414 2001 0.196 POINT (2.830552 42.15426)
# 9 EU2 85.97881 2001 0.196 POINT (-4.866867 39.29276)
# 10 EU2 41.99649 2005 0.196 POINT (-2.718447 42.86548)
#
# $all_plots
# Simple feature collection with 5328 features and 6 fields
# Geometry type: POINT
# Dimension: XY
# Bounding box: xmin: -7.451944 ymin: 36.06354 xmax: 3.24262 ymax: 43.46688
# Geodetic CRS: WGS 84
# First 10 features:
# PLOT_ID AGB_T_HA AVG_YEAR SIZE_HA geometry defo defo_start_year
# 299 EU2 45.90801 2002 0.196 POINT (-6.740698 38.14153) 0 NA
# 300 EU2 51.24531 2006 0.196 POINT (-5.06974 37.89271) 0 NA
# 301 EU2 69.61214 2004 0.196 POINT (-2.326055 38.2154) 0 NA
# 302 EU2 58.50748 2002 0.196 POINT (-4.500239 40.56834) 0 NA
# 303 EU2 130.73962 2000 0.196 POINT (-4.524572 43.13481) 0 NA
# 304 EU2 16.50173 2005 0.196 POINT (-0.9116234 42.72071) 0 NA
# 305 EU2 25.77160 2006 0.196 POINT (-5.17236 38.28734) 0 NA
# 306 EU2 77.99414 2001 0.196 POINT (2.830552 42.15426) 0 NA
# 307 EU2 85.97881 2001 0.196 POINT (-4.866867 39.29276) 0 NA
# 308 EU2 41.99649 2005 0.196 POINT (-2.718447 42.86548) 0 NAWe can see that the function returns a list containing two elements:
$non_deforested_plots: Asfobject with non-deforested plots.$all_plots: The original inputsfobject with added deforestated proportion (0-1) w.r.t. to plot area and deforestation start year.
In the next step we assign ecological zones and continents to plot locations. We use BiomePair() for this. This function overlays plot locations with pre-processed data to assign corresponding FAO ecological zones (biomes), Global Ecological Zones (GEZ) and continents (zones) to each plot:
preprocessed_plots_biome <- BiomePair(preprocessed_plots$non_deforested_plots)
head(preprocessed_plots_biome)
# PLOT_ID AGB_T_HA AVG_YEAR SIZE_HA POINT_X POINT_Y ZONE FAO.ecozone GEZ
# 1 EU2 26.57895 2001 0.196 1.967619 42.37488 Europe Temperate mountain system Temperate
# 2 EU2 207.19827 2001 0.196 1.943181 42.38366 Europe Temperate mountain system Temperate
# 3 EU2 106.25631 2001 0.196 2.319521 42.40453 Europe Temperate mountain system Temperate
# 4 EU2 56.08846 2001 0.196 1.820888 42.42750 Europe Temperate mountain system Temperate
# 5 EU2 143.96612 2001 0.196 3.036482 42.45157 Europe Subtropical dry forest Subtropical
# 6 EU2 100.97905 2001 0.196 1.723664 42.42646 Europe Temperate mountain system TemperateIn the step below we use TempApplyVar() to adjust plot biomass values to align with the map year by adding or subtracting annual growth increment to older or newer plots. The function uses growth data from a model-based estimate of growth-age relationships. The function also computes the temporal variance (standard deviation) of biomass changes based on growth-rate standard deviations. In the example below we apply temporal adjustment and calculate variance to all Global Ecological Zones (GEZ) within the plot data:
preprocessed_plots_biome_temp <- TempApplyVar(preprocessed_plots_biome, 2022)
head(preprocessed_plots_biome_temp)
# PLOT_ID AGB_T_HA AVG_YEAR SIZE_HA POINT_X POINT_Y ZONE FAO.ecozone GEZ AGB_T_HA_ORIG sdGrowth
# 1 EU2 91.67895 2001 0.196 1.967619 42.37488 Europe Temperate mountain system Temperate 26.57895 65.1
# 2 EU2 207.19827 2001 0.196 1.943181 42.38366 Europe Temperate mountain system Temperate 207.19827 0.0
# 3 EU2 106.25631 2001 0.196 2.319521 42.40453 Europe Temperate mountain system Temperate 106.25631 0.0
# 4 EU2 121.18846 2001 0.196 1.820888 42.42750 Europe Temperate mountain system Temperate 56.08846 65.1
# 5 EU2 164.96612 2001 0.196 3.036482 42.45157 Europe Subtropical dry forest Subtropical 143.96612 21.0
# 6 EU2 100.97905 2001 0.196 1.723664 42.42646 Europe Temperate mountain system Temperate 100.97905 0.0Note how the $AGB_T_HA, $AGB_T_HA_ORIG and $sdGrowth columns are added to the plot dataset. The first corresponding to the adjusted biomass values for the specified map_year, the second keeping the original biomass values before adjustment and the last adding the temporal standard deviation of the biomass adjustment.
AGB map validation
Now let’s suppose we want to validate an AGB map on the preprocessed_plots_biome_temp data above. We give an example of doing this with the invDasymetry() function where the AGB map is the ESA-CCI AGB dataset.
This function performs inverse dasymetric mapping on plot data. It selects plots based on given criteria, optionally aggregates them, and calculates tree cover fraction and AGB for each plot or cell. In our case we use a tree cover threshold of 10% meaning that only areas with tree cover percentage above this threshold will contribute to biomass estimates.
# Validate AGB map
AGBdata <- invDasymetry(plot_data = preprocessed_plots_biome_temp,
dataset = "esacci",
is_poly = FALSE,
threshold = 10,
map_year = 2022,
parallel = TRUE)
# memfrac : 0.5
# tolerance : 0.1
# verbose : FALSE
# todisk : FALSE
# tempdir : /tmp/RtmpjdE3bA
# datatype : FLT4S
# memmin : 1
# progress : 3
# 4975 plots or cells being processed...
# Processing 4975 plots in 100 batches of up to 50 plots each
# Processing complete. Results:
#
# plotAGB_10 tfPlotAGB orgPlotAGB mapAGB SIZE_HA x y
# 1 91.678952 91.67895 26.578952 49 0.196 1.9676189 42.37488
# 2 207.198266 207.19827 207.198266 95 0.196 1.9431806 42.38366
# 3 106.256307 106.25631 106.256307 133 0.196 2.3195210 42.40453
# 4 121.188457 121.18846 56.088457 6 0.196 1.8208879 42.42750
# 5 164.966116 164.96612 143.966116 105 0.196 3.0364816 42.45157
# 6 88.356669 100.97905 100.979051 114 0.196 1.7236641 42.42646
# (...)Now that we have the validation data from invDasymetry(), we can analyze and visualize the results. The function returns a data frame with several columns, including:
-
plotAGB_[threshold]: AGB values adjusted for forest cover threshold (e.g., plotAGB_10) -
tfPlotAGB: Tree-filtered plot AGB (only when not aggregated) -
orgPlotAGB: Original plot AGB values -
mapAGB: The AGB values extracted from the map -
SIZE_HA: Plot size in hectares -
xandy: Plot coordinates
We can now visualize the relationship between the reference plot data and map values using the plotting functions:
# Visualize results with binned plot
Binned(AGBdata$plotAGB_10, AGBdata$mapAGB,
"EU2 Plots - Binned Comparison")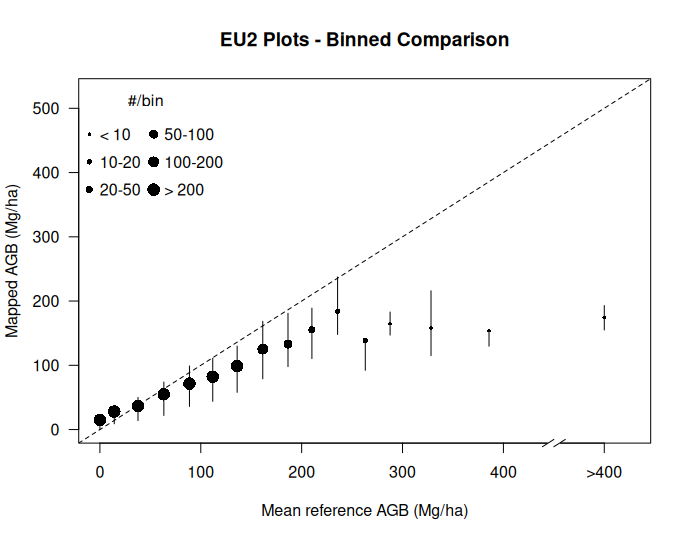
# Create a scatter plot
Scatter(AGBdata$plotAGB_10, AGBdata$mapAGB,
"EU2 Plots - Scatter Plot")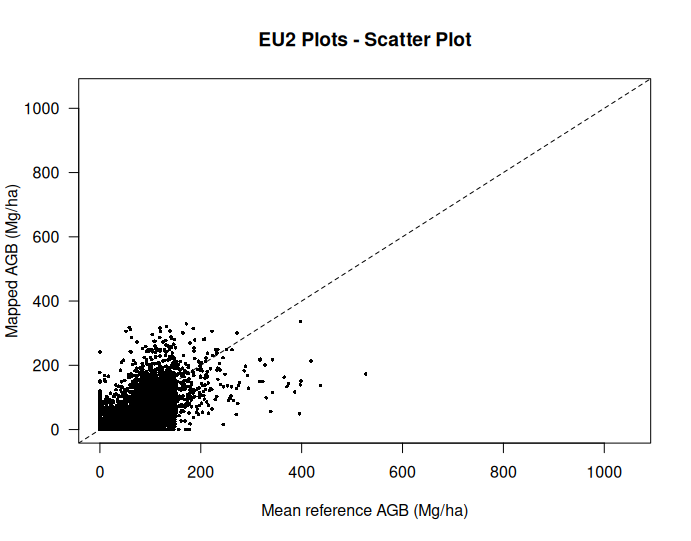
# Calculate accuracy metrics
# Use fewer intervals since we have a small sample
Accuracy(AGBdata, intervals = 6)
# AGB bin (Mg/ha) n AGBref (Mg/ha) AGBmap (Mg/ha) RMSD varPlot
# 1 0-100 3104 40 40 40 1
# 2 100-150 1556 121 89 63 1
# 3 150-200 157 170 128 76 1
# 4 200-250 53 219 166 84 1
# 5 250-300 19 270 146 139 1
# 6 >300 20 371 159 227 1
# 7 total 4909 74 60 53 1The output of the Accuracy() function provides an assessment of map accuracy including binned statistics by AGB ranges.
Handling Uncertainty
Plot2Map also provides a tool for calculating and incorporating plot uncertainty in plot data. The calculateTotalUncertainty() function combines measurement, sampling, and growth uncertainties. When uncertainty is not provided to plot data in the form of a varPlot column during inverse dasymetric mapping, invDasymetry() will call calculateTotalUncertainty() to calculate and add plot uncertainty to plot data automatically.
# Calculate total plot uncertainty
preprocessed_plots_biome_temp_uncertainty <- calculateTotalUncertainty(preprocessed_plots_biome_temp,
map_year = 2022,
map_resolution = 100)
# Calculating tree measurement uncertainty using RF model
# Calculating sampling uncertainty using Rejou-Mechain approach
# Loading sampling error data from package file
# Using existing growth uncertainty (sdGrowth) values
# Total uncertainty calculated for plot data of type: point
# Print the uncertainty components
print(preprocessed_plots_biome_temp_uncertainty$uncertainty_components)
# measurement sampling growth
# 0.2337400 0.1331422 0.6331177
# Inspect plot data with uncertainty
head(preprocessed_plots_biome_temp_uncertainty$data)
# PLOT_ID AGB_T_HA AVG_YEAR SIZE_HA POINT_X POINT_Y ZONE FAO.ecozone GEZ AGB_T_HA_ORIG sdGrowth sdTree RS_HA ratio sdSE varPlot sdTotal
# 1 EU2 91.67895 2001 0.196 1.967619 42.37488 Europe Temperate mountain system Temperate 26.57895 65.1 21.28590 1 0.196 22.01657 5175.8289 71.94323
# 2 EU2 207.19827 2001 0.196 1.943181 42.38366 Europe Temperate mountain system Temperate 207.19827 0.0 29.50758 1 0.196 22.01657 1355.4264 36.81612
# 3 EU2 106.25631 2001 0.196 2.319521 42.40453 Europe Temperate mountain system Temperate 106.25631 0.0 21.56276 1 0.196 22.01657 949.6819 30.81691
# 4 EU2 121.18846 2001 0.196 1.820888 42.42750 Europe Temperate mountain system Temperate 56.08846 65.1 24.19848 1 0.196 22.01657 5308.3057 72.85812
# 5 EU2 164.96612 2001 0.196 3.036482 42.45157 Europe Subtropical dry forest Subtropical 143.96612 21.0 35.91662 1 0.196 22.01657 2215.7332 47.07157
# 6 EU2 100.97905 2001 0.196 1.723664 42.42646 Europe Temperate mountain system Temperate 100.97905 0.0 21.25330 1 0.196 22.01657 936.4321 30.60118Aggregated Analysis
Plot2Map allows for spatial aggregation of plots into larger cells for analysis at different scales:
# Aggregated analysis at 0.1 degree resolution with minimum 3 plots per cell
AGBdata_agg <- invDasymetry(plot_data = preprocessed_plots_biome_temp_uncertainty$data,
clmn = "ZONE",
value = "Europe",
aggr = 0.1, # 0.1 degree aggregation
minPlots = 3, # Minimum 3 plots per cell
dataset = "esacci",
is_poly = FALSE)
# Visualize aggregated results
Binned(AGBdata_agg$plotAGB_10, AGBdata_agg$mapAGB,
"Europe - Aggregated 0.1 arc-deg")Working with Different Plot Types
Plot2Map can handle various types of plot data:
-
Point data with AGB estimates:
- Pre-formatted (see documentation)
- Unformatted (can be processed with
RawPlots())
-
Polygon data with corner coordinates:
- Can be processed with
Polygonize()
- Can be processed with
-
Tree-level measurement data:
- Processed with
MeasurementErr()which estimates AGB and uncertainty
- Processed with
-
Nested plot data (with sub-plots):
- Processed with
Nested()followed byMeasurementErr()
- Processed with
-
Lidar-based reference maps:
- Processed with
RefLidar()
- Processed with
See the “Plot data preparation” vignette for more information.
References
Araza et al., A comprehensive framework for assessing the accuracy and uncertainty of global above-ground biomass maps, Remote Sensing of Environment, Volume 272, 2022, 112917, ISSN 0034-4257, https://doi.org/10.1016/j.rse.2022.112917.
